Assessing the probability of metastatic mediastinal lymph node involvement in patients with non small cell lung cancer using convolutional neural networks on chest computed tomography
- Autores: Shevtsov A.E.1, Tominin I.D.1, Tominin V.D.1, Malevanniy V.M.1, Esakov Y.S.2, Tukvadze Z.G.2, Nefedov A.O.3, Yablonskii P.K.3, Gavrilov P.V.3, Kozlov V.V.4, Blokhina M.E.5, Nalivkina E.A.5, Gombolevskiy V.A.1,6, Vasilev Y.A.7, Dugova M.N.1, Chernina V.Y.1, Omelyanskaya O.V.7, Reshetnikov R.V.7, Blokhin I.A.7, Belyaev M.G.1
-
Afiliações:
- IRA Labs
- Moscow City Clinical Oncological Hospital № 1
- Saint-Petersburg State Research Institute of Phthisiopulmonology
- Novosibirsk Regional Clinical Oncology Dispensary
- AstraZeneca Pharmaceuticals LLC
- Artificial Intelligence Research Institute
- Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
- Edição: Volume 5, Nº 4 (2024)
- Páginas: 765-783
- Seção: Technical Reports
- URL: https://bakhtiniada.ru/DD/article/view/309835
- DOI: https://doi.org/10.17816/DD632008
- ID: 309835
Citar
Resumo
BACKGROUND: Lung cancer is the second most common cancer worldwide, accounting for approximately 20% of all cancer related deaths and having a <10% 5 year survival rate for very late stage cases. For the prevalent non small cell lung cancer (NSCLC), recent guidelines advise staging based on the 8th edition of the TNM classification, highlighting the importance of mediastinal lymph node involvement. While noninvasive methods are generally accurate, they often lack sensitivity, and invasive methods may not be suitable for all patients. Advances in deep learning present potential in solving such problems. However, most research focuses on algorithm development more than clinical relevance. Moreover, none of them addressed individual lymph node malignancies, limiting comprehensive analysis and interpretability and leaving clinicians without sufficient means to validate the results effectively.
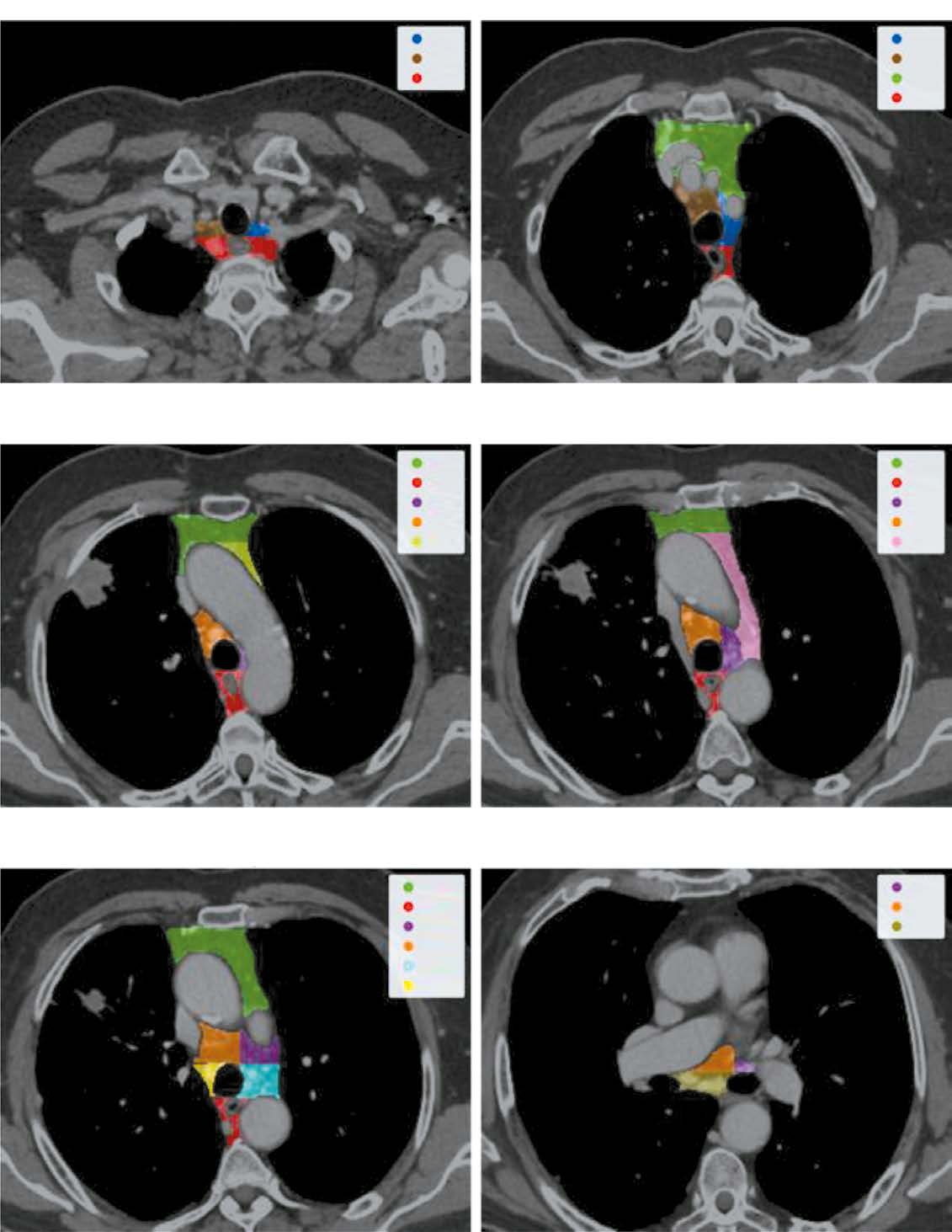
AIM: To develop a local data trained and validated algorithm for segmenting each mediastinal lymph node in chest computed tomography (CT) and assessing the probability of its involvement in metastasis.
MATERIALS AND METHODS: Initially, IASLC lymph node stations are segmented, providing a bounding box of the mediastinum for further processing. Next, the image is cropped to this box and passed through a second network to identify and mask all visible lymph nodes. Finally, each detected lymph node is extracted, stacked with its mask, and evaluated by a feed-forward network to determine malignancy probabilities.
RESULTS: The pipeline achieved an average recall and object Dice Score of 0.74±0.01 and 0.53±0.26 for the clinically relevant lymph node segmentation task. Further, it recorded a 0.73 ROC AUC for predicting a patient’s N-stage, outperforming traditional size based criteria.
CONCLUSIONS: The proposed algorithm enables new research algorithms to optimize the management of patients with nonenlarged intrathoracic lymph nodes, thus improving the quality of medical care for patients with cancer.
Palavras-chave
Texto integral
##article.viewOnOriginalSite##Sobre autores
Alexey Shevtsov
IRA Labs
Autor responsável pela correspondência
Email: a.shevtsov@ira-labs.com
ORCID ID: 0000-0003-3085-4325
Rússia, Moscow
Iaroslav Tominin
IRA Labs
Email: ya.tominin@ira-labs.com
ORCID ID: 0000-0002-7210-7208
Rússia, Moscow
Vladislav Tominin
IRA Labs
Email: v.tominin@ira-labs.com
ORCID ID: 0000-0001-5678-3452
Rússia, Moscow
Vsevolod Malevanniy
IRA Labs
Email: v.malevanniy@ira-labs.com
ORCID ID: 0009-0005-8804-2102
Rússia, Moscow
Yury Esakov
Moscow City Clinical Oncological Hospital № 1
Email: lungsurgery@mail.ru
ORCID ID: 0000-0002-5933-924X
Código SPIN: 8424-0756
MD, Cand. Sci. (Medicine)
Rússia, MoscowZurab Tukvadze
Moscow City Clinical Oncological Hospital № 1
Email: tukvadze.z.med@gmail.com
ORCID ID: 0000-0002-4550-6107
Rússia, Moscow
Andrey Nefedov
Saint-Petersburg State Research Institute of Phthisiopulmonology
Email: herurg78@mail.ru
ORCID ID: 0000-0001-6228-182X
Código SPIN: 2365-9458
MD, Cand. Sci. (Medicine)
Rússia, Saint PetersburgPiotr Yablonskii
Saint-Petersburg State Research Institute of Phthisiopulmonology
Email: glhirurgb2@mail.ru
ORCID ID: 0000-0003-4385-9643
Código SPIN: 3433-2624
MD, Dr. Sci. (Medicine), Professor
Rússia, Saint PetersburgPavel Gavrilov
Saint-Petersburg State Research Institute of Phthisiopulmonology
Email: spbniifrentgen@mail.ru
ORCID ID: 0000-0003-3251-4084
Código SPIN: 7824-5374
MD, Cand. Sci. (Med.)
Rússia, Saint PetersburgVadim Kozlov
Novosibirsk Regional Clinical Oncology Dispensary
Email: vadimkozlov80@mail.ru
ORCID ID: 0000-0003-3211-5139
Código SPIN: 8045-4286
MD, Cand. Sci. (Medicine)
Rússia, NovosibirskMariya Blokhina
AstraZeneca Pharmaceuticals LLC
Email: mariya.blokhina@astrazeneca.com
ORCID ID: 0009-0002-9008-9485
MD
Rússia, MoscowElena Nalivkina
AstraZeneca Pharmaceuticals LLC
Email: elena.nalivkina@astrazeneca.com
ORCID ID: 0009-0003-5412-9643
Rússia, Moscow
Victor Gombolevskiy
IRA Labs; Artificial Intelligence Research Institute
Email: gombolevskii@gmail.com
ORCID ID: 0000-0003-1816-1315
Código SPIN: 6810-3279
MD, Cand. Sci. (Med.)
Rússia, Moscow; MoscowYuriy Vasilev
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: VasilevYA1@zdrav.mos.ru
ORCID ID: 0000-0002-5283-5961
Código SPIN: 4458-5608
MD, Dr. Sci. (Medicine)
Rússia, MoscowMariya Dugova
IRA Labs
Email: m.dugova@ira-labs.com
ORCID ID: 0009-0004-5586-8015
MD
Rússia, MoscowValeria Chernina
IRA Labs
Email: v.chernina@ira-labs.com
ORCID ID: 0000-0002-0302-293X
Código SPIN: 8896-8051
MD
Rússia, MoscowOlga Omelyanskaya
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: OmelyanskayaOV@zdrav.mos.ru
ORCID ID: 0000-0002-0245-4431
Código SPIN: 8948-6152
Rússia, Moscow
Roman Reshetnikov
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: reshetnikov@fbb.msu.ru
ORCID ID: 0000-0002-9661-0254
Código SPIN: 8592-0558
Cand. Sci. (Physics and Mathematics)
Rússia, MoscowIvan Blokhin
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: BlokhinIA@zdrav.mos.ru
ORCID ID: 0000-0002-2681-9378
Código SPIN: 3306-1387
MD, Cand. Sci. (Medicine)
Rússia, MoscowMikhail Belyaev
IRA Labs
Email: belyaevmichel@gmail.com
ORCID ID: 0000-0001-9906-6453
Código SPIN: 2406-1772
Cand. Sci. (Physics and Mathematics)
Rússia, MoscowBibliografia
- Thandra KCh, Barsouk A, Saginala K, et al. Epidemiology of lung cancer. Contemporary Oncology. 2021;25(1):45–52. doi: 10.5114/wo.2021.103829
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. doi: 10.1016/j.jtho.2015.09.009
- Tanoue LT, Tanner NT, Gould MK, Silvestri GA. Lung cancer screening. Am J Respir Crit Care Med. 2015;191(1):19–33. doi: 10.1164/rccm.201410-1777CI
- Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: Non small cell lung cancer, version 1. 2020. J Natl Compr Canc Netw. 2019;17(12):1464–1472. doi: 10.6004/jnccn.2019.0059
- Planchard D, Popat S, Kerr K, et al. Metastatic non small cell lung cancer: ESMO clin-ical practice guidelines for diagnosis, treatment and follow up [published correction appears in Ann Oncol. 2019;30(5):863–870. doi: 10.1093/annonc/mdy474]. Ann Oncol. 2018;29(Suppl 4):iv192–iv237. doi: 10.1093/annonc/mdy275
- Heleno B, Siersma V, Brodersen J. Estimation of overdiagnosis of lung cancer in low dose computed tomography screening: A secondary analysis of the danish lung cancer screening trial. JAMA Intern Med. 2018;178(10):1420–1422. doi: 10.1001/jamainternmed.2018.3056
- Lopes Pegna A, Picozzi G, Falaschi F, et al. Four year results of low dose CT screen ing and nodule management in the ITALUNG trial. J Thorac Oncol. 2013;8(7):866–875. doi: 10.1097/JTO.0b013e31828f68d6
- Infante M, Cavuto S, Lutman FR, et al. Long term follow up results of the DANTE trial, a randomized study of lung cancer screening with spiral computed tomography. Am J Respir Crit Care Med. 2015;191(10):1166–1175. doi: 10.1164/rccm.201408-1475OC
- De Koning H, van der Aalst C, de Jong P. Reduced lung cancer mortality with volume CT screening in a randomized trial. N Engl J Med. 2020;382(6):503–513. doi: 10.1056/NEJMoa1911793
- Pastorino U, Silva M, Sestini S, et al. Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: new confirmation of lung cancer screening efficacy. Ann Oncol. 2019;30(10):1672. doi: 10.1093/annonc/mdz169
- Baldwin DR, Duffy SW, Wald NJ, et al. UK Lung Screen (UKLS) nodule management protocol: modelling of a single screen randomised controlled trial of low dose CT screening for lung cancer. Thorax. 2011;66(4):308–313. doi: 10.1136/thx.2010.152066
- Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151(1):193–203. doi: 10.1016/j.chest.2016.10.010
- Nakajima T, Yasufuku K, Yoshino I. Current status and perspective of EBUS-TBNA. Gen Thorac Cardiovasc Surg. 2013;61(7):390–396. doi: 10.1007/s11748-013-0224-6
- Hartert M, Tripsky J, Huertgen M. Video-assisted mediastinoscopic lymphadenecto-my (VAMLA) for staging & treatment of non small cell lung cancer (NSCLC). Mediastinum. 2020;4:3. doi: 10.21037/med.2019.09.06
- Ettinger DS, Wood DE, Aisner DL, et al. Non small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050
- Roberts PF, Follette DM, von Haag D, et al. Factors associated with false positive staging of lung cancer by positron emission tomography. Ann Thorac Surg. 2000;70(4):1154–1160. doi: 10.1016/s0003-4975(00)01769-0
- Kanzaki R, Higashiyama M, Fujiwara A, et al. Occult mediastinal lymph node metas-tasis in NSCLC patients diagnosed as clinical N0-1 by preoperative integrated FDG-PET/CT and CT: risk factors, pattern, and histopathological study. Lung Cancer. 2011;71(3):333–337. doi: 10.1016/j.lungcan.2010.06.008
- Verduzco-Aguirre HC, Lopes G, Soto Perez De Celis E. Implementation of diagnostic resources for cancer in developing countries: a focus on PET/CT. Ecancermedical science. 2019;13:ed87. doi: 10.3332/ecancer.2019.ed87
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539
- Guo D, Ye X, Ge J, et al. Deepstationing: thoracic lymph node station parsing in CT scans using anatomical context encoding and key organ auto search. In: International Conference on Medical Image Computing and Computer Assisted Intervention; 2021 September 27–October 1; Strasbourg. Available from: https://miccai2021.org/openaccess/paperlinks/2021/09/01/140-Paper0015.html
- Iuga AI, Carolus H, Höink AJ, et al. Automated detection and segmentation of thorac ic lymph nodes from CT using 3D foveal fully convolutional neural networks. BMC Med Imaging. 2021;21(1):69. doi: 10.1186/s12880-021-00599-z
- Iuga AI, Lossau T, Caldeira LL, et al. Automated mapping and N-staging of thoracic lymph nodes in contrast-enhanced CT scans of the chest using a fully convolutional neural network. Eur J Radiol. 2021;139:109718. doi: 10.1016/j.ejrad.2021.109718
- Zhong Y, Yuan M, Zhang T, et al. Radiomics approach to prediction of occult medi astinal lymph node metastasis of lung adenocarcinoma. AJR Am J Roentgenol. 2018;211(1):109–113. doi: 10.2214/AJR.17.19074
- Liu Y, Kim J, Balagurunathan Y, et al. Prediction of pathological nodal involvement by CT-based Radiomic features of the primary tumor in patients with clinically node negative pe ripheral lung adenocarcinomas. Med Phys. 2018;45(6):2518–2526. doi: 10.1002/mp.12901
- Cong M, Yao H, Liu H, et al. Development and evaluation of a venous computed to mography radiomics model to predict lymph node metastasis from non small cell lung cancer. Medicine (Baltimore). 2020;99(18):e20074. doi: 10.1097/MD.0000000000020074
- Gu P, Zhao YZ, Jiang LY, et al. Endobronchial ultrasound guided transbronchial nee dle aspiration for staging of lung cancer: a systematic review and meta analysis. Eur J Cancer. 2009;45(8):1389–1396. doi: 10.1016/j.ejca.2008.11.043
- Brown G, Richards CJ, Bourne MW, et al. Morphologic predictors of lymph node sta tus in rectal cancer with use of high spatial resolution MR imaging with histopathologic comparison. Radiology. 2003;227(2):371–377. doi: 10.1148/radiol.2272011747
- Som PM. Lymph nodes of the neck. Radiology. 1987;165(3):593–600. doi: 10.1148/radiology.165.3.3317494
- Curtin HD, Ishwaran H, Mancuso AA, et al. Comparison of CT and MR imaging in staging of neck metastases. Radiology. 1998;207(1):123–130. doi: 10.1148/radiology.207.1.9530307
- Loch FN, Asbach P, Haas M, et al. Accuracy of various criteria for lymph node stag ing in ductal adenocarcinoma of the pancreatic head by computed tomography and magnetic reso nance imaging. World J Surg Oncol. 2020;18(1):213. doi: 10.1186/s12957-020-01951-3
- Elsholtz FH, Asbach P, Haas M, et al. Introducing the node reporting and data system 1.0 (Node-RADS): a concept for standardized assessment of lymph nodes in cancer Eur Radiol. 2021;31(8):7217. Eur Radiol. 2021;31(9):6116–6124. doi: 10.1007/s00330-020-07572-4 Сorrected and republished from: Eur Radiol. 2021;31(9): 7217. doi: 10.1007/s00330-021-07795-z
- Ceylan N, Doğan S, Kocaçelebi K, et al. Contrast enhanced CT versus integrated PET-CT in pre-operative nodal staging of non-small cell lung cancer. Diagn Interv Radiol. 2012;18(5):435–440. doi: 10.4261/1305-3825.DIR.5100-11.2
- Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004
- Çiçek Ö, Abdulkadir A, Lienkamp SS, et al. 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer Assisted Inter vention (MICCAI 2016), Part II: 19th International Conference; 2016 October 17–21; Athens. P. 424–432.
- Milletari F, Navab N, Ahmadi SA. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth international conference on 3D vision (3DV): proceedings article. 2016 October 25–28; California. P. 565–571. doi: 10.1109/3DV.2016.79
- Van Ginneken B, Armato SG, de Hoop B, et al. Comparing and combining algorithms for computer aided detection of pulmonary nodules in computed tomography scans: the ANODE09 study. Med Image Anal. 2010;14(6):707–722. doi: 10.1016/j.media.2010.05.005
- Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. The international multimodal brain tumor segmentation (BraTS) challenge. 2018. doi: 10.48550/arXiv.1811.02629
- Silva F, Pereira T, Frade J, et al. Pre training autoencoder for lung nodule malignancy assessment using CT images. Applied Sciences. 2020;10(21):7837. doi: 10.3390/app10217837
- Dubost F, Adams H, Yilmaz P, et al. Weakly supervised object detection with 2D and 3D regression neural networks. Med Image Anal. 2020;65:101767. doi: 10.1016/j.media.2020.101767
- Rusch VW, Asamura H, Watanabe H, et al. The IASLC lung cancer staging project: a proposal for a new international lymph node map in the forthcoming seventh edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4(5):568–577. doi: 10.1097/JTO.0b013e3181a0d82e
- Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer Assisted Intervention (MICCAI 2015): 18th International Conference; 2015 May; Munich; Р. 234–241. doi: 10.48550/arXiv.1505.04597
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2016 June 27–30; Las Vegas. P. 770–778. doi: 10.48550/arXiv.1512.03385
- Ioffe S, Szegedy Ch. Batch normalization: accelerating deep network training by reducing internal covariate shift. ArXiv. 2015;1. doi: 10.48550/arXiv.1502.03167
- Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. In: Conference: proceedings of the 27th International Conference on Machine Learning (ICML-10); 2010 June 21–24; Haifa. Available from: https://icml.cc/Conferences/2010/papers/432.pdf
- Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. In: Conference: proceedings of the 27th International Conference on Machine Learning (ICML-10), June 21–24, 2010. Haifa, Israel; 2010. Р. 807–814.
- Roth HR, Lu L, Seff A, et al. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. Med Image Comput Comput Assist Interv. 2014;17(1):520–527. doi: 10.1007/978-3-319-10404-1_65
- Goncharov M, Pisov M, Shevtsov A, et al. CT-based COVID-19 triage: deep multi-task learning improves joint identification and severity quantification. Med Image Anal. 2021;71:102054. doi: 10.1016/j.media.2021.102054
Arquivos suplementares