Application of large language models in radiological diagnostics: a scoping review
- Authors: Vasilev Y.A.1, Reshetnikov R.V.1, Nanova O.G.1, Vladzymyrskyy A.V.1, Arzamasov K.M.1, Omelyanskaya O.V.1, Kodenko M.R.1, Erizhokov R.A.1, Pamova A.P.1, Seradzhi S.R.1, Blokhin I.A.1, Gonchar A.P.1,2, Gelezhe P.B.1, Akhmedzyanova D.A.1, Shumskaya Y.F.1
-
Affiliations:
- Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
- Moscow City Hospital named after S.S. Yudin
- Issue: Vol 6, No 2 (2025)
- Pages: 268-285
- Section: Systematic reviews
- URL: https://bakhtiniada.ru/DD/article/view/310215
- DOI: https://doi.org/10.17816/DD678373
- EDN: https://elibrary.ru/QSANCA
- ID: 310215
Cite item
Full Text
Abstract
BACKGROUND: Modern large language models show potential for application in radiological diagnostics across a wide range of routine tasks.
AIM: The work aimed to conduct a scoping review of the application of large language models in radiological diagnostics by analyzing possible use-case scenarios and assessing the methodological quality of relevant studies.
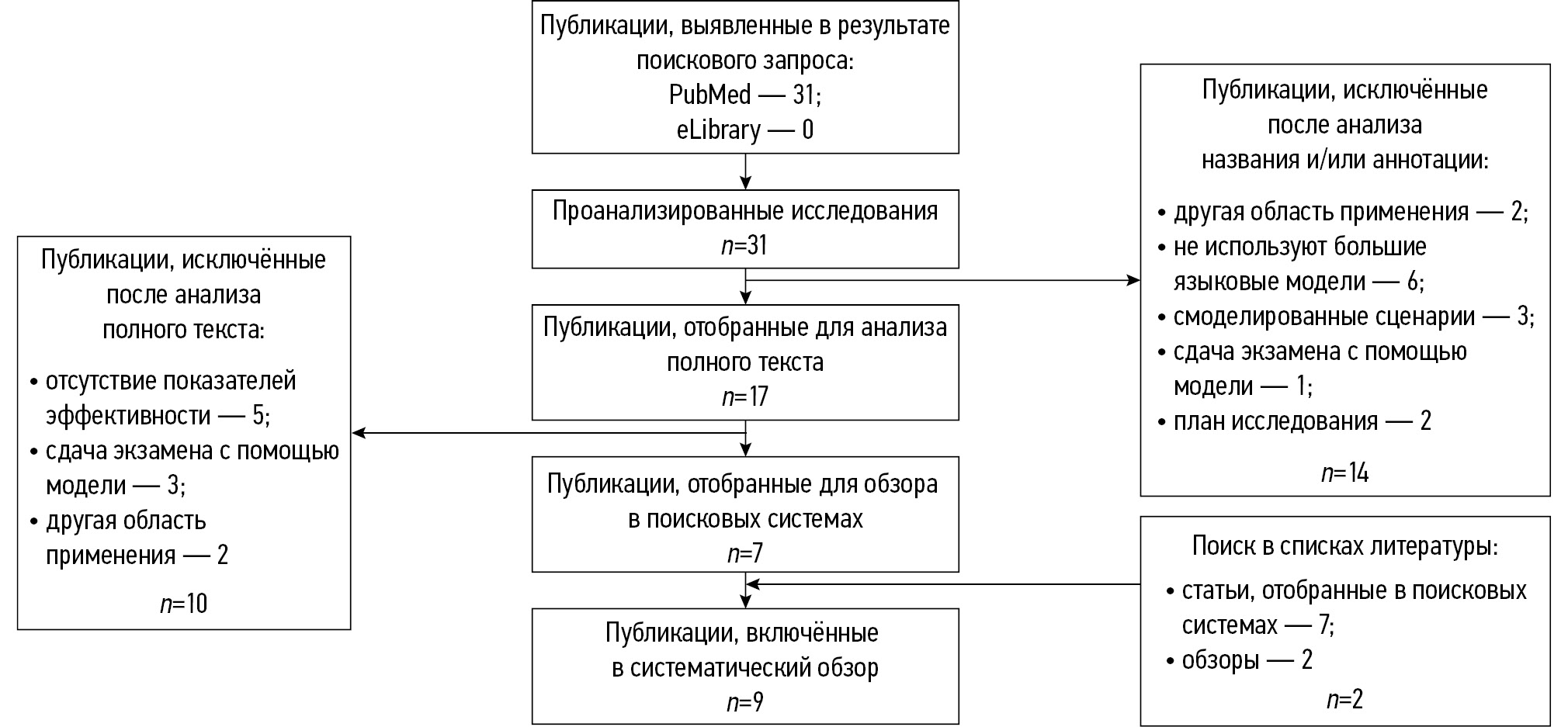
METHODS: Two search strategies were employed: a primary search (PubMed and eLibrary) targeting full-text publications with well-developed methodology, and a supplementary search (PubMed) aimed at broader coverage of large language model use cases in radiological diagnostics during 2023–2025. Extracted data included bibliometric characteristics, study objectives, use-case scenarios of large language models, nosological profiles, key methodological parameters, and both quantitative and qualitative indicators of diagnostic performance—for both the models and the specialists involved, including their number and experience. The quality was assessed using the modified QUADAS-CAD questionnaire.
RESULTS: The primary search yielded 9 studies for analysis; the supplementary search yielded 216. A total of 9 major use-case scenarios for large language models in radiology were identified. The most common among them was the rephrasing of radiology reports in order to improve their accessibility for patient understanding. Models predominantly used were GPT-4 and BERT, along with GPT-3.5, Llama 2, Med42, GPT-4V, and Gemini Pro. The large language model GPT-4 demonstrated high diagnostic accuracy in identifying brain tumors (73.0%), myocarditis (83.0%), and in making decisions on invasive procedures for acute coronary syndrome (86.0%). In turn, it demonstrated low diagnostic accuracy for nervous system disorders of various etiologies (50.0%) and for musculoskeletal diseases (43.0%). The BERT model exhibited high diagnostic accuracy in detecting pulmonary nodules (99.0%) and signs of intracranial hemorrhage (sensitivity and specificity: 97.0% and 90.0%, respectively), as well as in report classification (accuracy: 84.3%).
Most articles (88.9%) carried a high risk of bias. The main reasons for this included small and imbalanced sample sizes, overlap between training and test datasets, and insufficiently precise preparation and description of reference standards.
CONCLUSION: The diagnostic performance of large language models varies significantly across articles. Their clinical implementation requires standardized, methodologically robust research, including larger and more balanced samples, optimization of the structure and volume of datasets, separation of training and testing samples, thorough preparation and description of reference standards, as well as the accumulation of empirical data for specific radiological tasks.
Full Text
##article.viewOnOriginalSite##About the authors
Yuriy A. Vasilev
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: npcmr@zdrav.mos.ru
ORCID iD: 0000-0002-5283-5961
SPIN-code: 4458-5608
MD, Cand. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Roman V. Reshetnikov
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: ReshetnikovRV1@zdrav.mos.ru
ORCID iD: 0000-0002-9661-0254
SPIN-code: 8592-0558
Cand. Sci. (Physics and Mathematics)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Olga G. Nanova
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Author for correspondence.
Email: nanova@mail.ru
ORCID iD: 0000-0001-8886-3684
SPIN-code: 6135-4872
Cand. Sci. (Biology)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Anton V. Vladzymyrskyy
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: VladzimirskijAV@zdrav.mos.ru
ORCID iD: 0000-0002-2990-7736
SPIN-code: 3602-7120
MD, Dr. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Kirill M. Arzamasov
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: ArzamasovKM@zdrav.mos.ru
ORCID iD: 0000-0001-7786-0349
SPIN-code: 3160-8062
MD, Dr. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Olga V. Omelyanskaya
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: o.omelyanskaya@npcmr.ru
ORCID iD: 0000-0002-0245-4431
SPIN-code: 8948-6152
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051
Maria R. Kodenko
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: KodenkoMR@zdrav.mos.ru
ORCID iD: 0000-0002-0166-3768
SPIN-code: 5789-0319
Cand. Sci. (Engineering)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Rustam A. Erizhokov
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: ErizhokovRA@zdrav.mos.ru
ORCID iD: 0009-0007-3636-2889
SPIN-code: 2274-6428
MD
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Anastasia P. Pamova
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: PamovaAP@zdrav.mos.ru
ORCID iD: 0000-0002-0041-3281
SPIN-code: 5146-4355
MD, Cand. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Seal R. Seradzhi
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: SeradzhiSR@zdrav.mos.ru
ORCID iD: 0009-0000-3990-6668
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051
Ivan A. Blokhin
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: BlokhinIA@zdrav.mos.ru
ORCID iD: 0000-0002-2681-9378
SPIN-code: 3306-1387
MD, Cand. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Anna P. Gonchar
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies; Moscow City Hospital named after S.S. Yudin
Email: GoncharAP@zdrav.mos.ru
ORCID iD: 0000-0001-5161-6540
SPIN-code: 3513-9531
MD, Cand. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051; MoscowPavel B. Gelezhe
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: GelezhePB@zdrav.mos.ru
ORCID iD: 0000-0003-1072-2202
SPIN-code: 4841-3234
MD, Cand. Sci. (Medicine)
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Dina A. Akhmedzyanova
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: AkhmedzyanovaDA@zdrav.mos.ru
ORCID iD: 0000-0001-7705-9754
SPIN-code: 6983-5991
MD
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051Yuliya F. Shumskaya
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: shumskayayf@zdrav.mos.ru
ORCID iD: 0000-0002-8521-4045
SPIN-code: 3164-5518
MD
Russian Federation, 24 Petrovka st, bldg 1, Moscow, 127051References
- Cherif H, Moussa C, Missaoui AM, et al. Appraisal of ChatGPT’s aptitude for medical education: comparative analysis with third-year medical students in a pulmonology examination. JMIR Medical Education. 2024;10:e52818. doi: 10.2196/52818 EDN: OFMTDE
- Kim W, Kim BC, Yeom HG. Performance of large language models on the Korean Dental licensing examination: a comparative study. International Dental Journal. 2025;75(1):176–184. doi: 10.1016/j.identj.2024.09.002 EDN: JDFMDL
- Busch F, Hoffmann L, dos Santos DP, et al. Large language models for structured reporting in radiology: past, present, and future. European Radiology. 2024;35(5):2589–2602. doi: 10.1007/s00330-024-11107-6 EDN: PNFKNR
- Lecler A, Duron L, Soyer P. Revolutionizing radiology with GPT-based models: Current applications, future possibilities and limitations of ChatGPT. Diagnostic and Interventional Imaging. 2023;104(6):269–274. doi: 10.1016/j.diii.2023.02.003EDN: FGMMTY
- Tricco AC, Lillie E, Zarin W, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Annals of Internal Medicine. 2018;169(7):467–473. doi: 10.7326/M18-0850
- Vasilev YuA, Vladzymyrskyy AV, Omelyanskaya OV, et al. Methodological recommendations for preparing a systematic review. Moscow: Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies; 2023. (In Russ.) EDN: XKXHDA
- Kodenko MR, Vasilev YA, Vladzymyrskyy AV, et al. Diagnostic accuracy of ai for opportunistic screening of abdominal aortic aneurysm in CT: a systematic review and narrative synthesis. Diagnostics. 2022;12(12):3197. doi: 10.3390/diagnostics12123197 EDN: ERWYPX
- Horiuchi D, Tatekawa H, Oura T, et al. ChatGPT’s diagnostic performance based on textual vs. visual information compared to radiologists’ diagnostic performance in musculoskeletal radiology. European Radiology. 2024;35(1):506–516. doi: 10.1007/s00330-024-10902-5 EDN: JAHWFM
- Mitsuyama Y, Tatekawa H, Takita H, et al. Comparative analysis of GPT-4-based ChatGPT’s diagnostic performance with radiologists using real-world radiology reports of brain tumors. European Radiology. 2024;35(4):1938–1947. doi: 10.1007/s00330-024-11032-8 EDN: UHMLBQ
- Kaya K, Gietzen C, Hahnfeldt R, et al. Generative Pre-trained Transformer 4 analysis of cardiovascular magnetic resonance reports in suspected myocarditis: A multicenter study. Journal of Cardiovascular Magnetic Resonance. 2024;26(2):101068. doi: 10.1016/j.jocmr.2024.101068 EDN: TSRLJX
- Grolleau E, Couraud S, Jupin Delevaux E, et al. Incidental pulmonary nodules: Natural language processing analysis of radiology reports. Respiratory Medicine and Research. 2024;86:101136. doi: 10.1016/j.resmer.2024.101136 EDN: DHDPIX
- Khoruzhaya AN, Kozlov DV, Arzamasov KM, Kremneva EI. Comparison of an ensemble of machine learning models and the BERT language model for analysis of text descriptions of brain CT reports to determine the presence of intracranial hemorrhage. Sovremennye tehnologii v medicine. 2024;16(1):27–36. doi: 10.17691/stm2024.16.1.03 EDN: AXXVVD
- Han T, Adams LC, Bressem KK, et al. Comparative analysis of multimodal large language model performance on clinical vignette questions. JAMA. 2024;331(15):1320–1321. doi: 10.1001/jama.2023.27861 EDN: KPFLZG
- Horiuchi D, Tatekawa H, Shimono T, et al. Accuracy of ChatGPT generated diagnosis from patient's medical history and imaging findings in neuroradiology cases. Neuroradiology. 2023;66(1):73–79. doi: 10.1007/s00234-023-03252-4 EDN: SRFGAA
- Wataya T, Miura A, Sakisuka T, et al. Comparison of natural language processing algorithms in assessing the importance of head computed tomography reports written in Japanese. Japanese Journal of Radiology. 2024;42(7):697–708. doi: 10.1007/s11604-024-01549-9 EDN: VAKPBV
- Cagnina A, Salihu A, Meier D, et al. Assessing the need for coronary angiography in high-risk non-ST-elevation acute coronary syndrome patients using artificial intelligence and computed tomography. The International Journal of Cardiovascular Imaging. 2024;41(1):55–61. doi: 10.1007/s10554-024-03283-9 EDN: JMBFSX
- Gallifant J, Afshar M, Ameen S, et al. The TRIPOD-LLM reporting guideline for studies using large language models. Nature Medicine. 2025;31(1):60–69. doi: 10.1038/s41591-024-03425-5 EDN: KAPIXF
- Tripathi S, Alkhulaifat D, Doo FX, et al. Development, evaluation, and assessment of large language models (DEAL) checklist: a technical report. NEJM AI. 2025;2(6). doi: 10.1056/AIp2401106
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B: Statistical Methodology. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
- Hollestein LM, Lo SN, Leonardi-Bee J, et al. MULTIPLE ways to correct for MULTIPLE comparisons in MULTIPLE types of studies. British Journal of Dermatology. 2021;185(6):1081–1083. doi: 10.1111/bjd.20600 EDN: QQWVVP
- Collins GS, Moons KGM, Dhiman P, et al. TRIPOD+AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ. 2024;385:e078378. doi: 10.1136/bmj-2023-078378 EDN: WSTQKK
- Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6(11):e012799. doi: 10.1136/bmjopen-2016-012799
- Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527
- Vasiliev YuA, Vlazimirsky AV, Omelyanskaya OV, et al. Methodology for testing and monitoring artificial intelligence-based software for medical diagnostics. Digital Diagnostics. 2023;4(3):252–267. doi: 10.17816/DD321971 EDN: UEDORU
- Vasilev YuA, Bobrovskaya TM, Arzamasov KM, et al. Medical datasets for machine learning: fundamental principles of standartization and systematization. Manager Zdravookhranenia. 2023; (4):28–41. doi: 10.21045/1811-0185-2023-4-28-41 EDN: EPGAMD
- Vinogradova IA, Nizovtsova LA, Omelyanskaya OV. Innovative strategic session in the scientific activity of the Center for Diagnostics and Telemedicine. Digital Diagnostics. 2022;3(4):414–420. doi: 10.17816/DD111833 EDN: DLRLVI
- Kalinina ML, Svitachev AP, Biswas D, Vishnu P. Comparison of awareness and attitudes toward artificial intelligence among Russian- and English-speaking students at Orenburg State Medical University. Digital Diagnostics. 2023;4(1S):62–65. doi: 10.17816/DD430346 EDN: DIKOYA
Supplementary files